Get Started - Tutorial¶
Pyevolve combined with Python language can be a powerful tool. The best way to show you how Pyevolve can be used, is beginning with simple examples, later we’ll show some snippets and etc. So you’ll can walk by yourself.
First Example¶
To make the API easy to use, we have created default parameters for almost every parameter in Pyevolve, for example, when you will use the G1DList.G1DList genome without specifying the Mutator, Crossover and Initializator, you will use the default ones: Swap Mutator, One Point Crossover and the Integer Initialzator. All those default parameters are specified in the Consts module (and you are highly encouraged to take a look at source code).
Let’s begin with the first simple example (Ex. 1). First of all, you must know your problem, in this case, our problem is to find a simple 1D list of integers of n-size with zero in all positions. At the first look, we know by intuition that the representation needed to this problem is a 1D List, which you can found in Pyevolve by the name of G1DList.G1DList, which means Genome 1D List. This representation is based on a python list as you will see, and is very easy to manipulate. The next step is to define the our evaluation function to our Genetic Algorithm. We want all the n list positions with value of ‘0’, so we can propose the evaluation function:
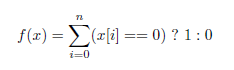
As you can see in the above equation, with the x variable representing our genome list of integers, the f(x) shows our evaluation function, which is the sum of ‘0’ values in the list. For example, if we have a list with 10 elements like this:
x = [1, 2, 3, 8, 0, 2, 0, 4, 1, 0]
we will got the raw score [1] value of 3, or f(x) = 3. It’s very simple to understand. Now, let’s code this.
We will define our evaluation function “eval_func” as:
# This function is the evaluation function, we want
# to give high score to more zero'ed chromosomes
def eval_func(chromosome):
score = 0.0
# iterate over the chromosome elements (items)
for value in chromosome:
if value==0:
score += 1.0
return score
As you can see, this evaluation function verify each element in the list which is equal to ‘0’ and return the proportional score value. The G1DList.G1DList chromosome is not a python list by itself but it encapsulates one and exposes the methods for this list, like the iterator used in the above loop. The next step is the creation of an one sample genome [2] for the Genetic Algorithm. We can define our genome as this:
# Genome instance
genome = G1DList.G1DList(20)
# The evaluator function (objective function)
genome.evaluator.set(eval_func)
This will create an instance of the G1DList.G1DList class (which resides in the G1DList module) with the list n-size of 20 and sets the evaluation function of the genome to the evaluation function “eval_func” that we have created before.
But wait, where is the range of integers that will be used in the list ? Where is the mutator, crossover and initialization functions ? They are all in the default parameters, as you see, this parameters keep things simple.
By default (and you have the documentation to find this defaults), the range of the integers in the G1DList.G1DList is between the inverval [ Consts.CDefRangeMin, Consts.CDefRangeMax] inclusive, and genetic operators is the same I have cited before: Swap Mutator Mutators.G1DListMutatorSwap(), One Point Crossover Crossovers.G1DListCrossoverSinglePoint() and the Integer Initializator Initializators.G1DListInitializatorInteger(). You can change everything with the API, for example, you can pass the ranges to the genome, like this:
genome.setParams(rangemin=0, rangemax=10)
Right, now we have our evaluation function and our first genome ready, the next step is to create our Genetic Algorithm Engine, the GA Core which will do the evolution, control statistics, etc... The GA Engine which we will use is the GSimpleGA.GSimpleGA which resides in the GSimpleGA module, this GA Engine is the genetic algorithm [3] described by Goldberg. So, let’s create the engine:
ga = GSimpleGA.GSimpleGA(genome)
Ready ! Simple not ? We simple create our GA Engine with the created genome. You can ask: “Where is the selector method ? The number of generations ? Mutation rate ?“. Again: we have defaults. By default, the GA will evolve for 100 generations with a population size of 80 individuals, it will use the mutation rate of 2% and a crossover rate of 80%, the default selector is the Ranking Selection (Selectors.GRankSelector()) method. Those default parameters was not random picked, they are all based on the commom used properties.
Now, all we need to do is to evolve !
# Do the evolution, with stats dump
# frequency of 10 generations
ga.evolve(freq_stats=10)
# Best individual
print ga.bestIndividual()
Note
Pyevolve have the __repr__() function implemented for almost all objects, this means that you can use syntax like ‘print object’ and the object information will be show in an pretty format.
Ready, now we have our first Genetic Algorithm, it looks more like a “Hello GA !” application. The code above shows the call of the GSimpleGA.GSimpleGA.evolve() method, with the parameter freq_stats=10, this method will do the evolution and will show the statistics every 10th generation; the next method called is the GSimpleGA.GSimpleGA.bestIndividual(), this method will return the best individual after the end of the evolution, and the with the print python command, we will show the genome on the screen.
This is what this example will results:
Gen. 1 (1.00%): Max/Min/Avg Fitness(Raw) [2.40(3.00) / 1.60(1.00) / 2.00(2.00)]
Gen. 10 (10.00%): Max/Min/Avg Fitness(Raw) [10.80(10.00) / 7.20(8.00) / 9.00(9.00)]
Gen. 20 (20.00%): Max/Min/Avg Fitness(Raw) [22.80(20.00) / 15.20(18.00) / 19.00(19.00)]
Gen. 30 (30.00%): Max/Min/Avg Fitness(Raw) [20.00(20.00) / 20.00(20.00) / 20.00(20.00)]
(...)
Gen. 100 (100.00%): Max/Min/Avg Fitness(Raw) [20.00(20.00) / 20.00(20.00) / 20.00(20.00)]
Total time elapsed: 3.375 seconds.
- GenomeBase
Score: 20.000000
Fitness: 20.000000
Slot [Evaluator] (Count: 1)
Name: eval_func
Slot [Initializator] (Count: 1)
Name: G1DListInitializatorInteger
Doc: Integer initialization function of G1DList,
accepts 'rangemin' and 'rangemax'
Slot [Mutator] (Count: 1)
Name: G1DListMutatorSwap
Doc: The mutator of G1DList, Swap Mutator
Slot [Crossover] (Count: 1)
Name: G1DListCrossoverSinglePoint
Doc: The crossover of G1DList, Single Point
- G1DList
List size: 20
List: [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]
This is the evolution of our Genetic Algorithm with the best individual show at the end of the evolution. As you can see, the population have obtained the best raw score (20.00) near the generation 20.
Final source code¶
Here is the final source code:
from pyevolve import G1DList
from pyevolve import GSimpleGA
def eval_func(chromosome):
score = 0.0
# iterate over the chromosome
for value in chromosome:
if value==0:
score += 1
return score
genome = G1DList.G1DList(20)
genome.evaluator.set(eval_func)
ga = GSimpleGA.GSimpleGA(genome)
ga.evolve(freq_stats=10)
print ga.bestIndividual()
Footnotes
| [1] | It is important to note that in Pyevolve, we have raw score and fitness score, the raw score is the return of the evaluation function and the fitness score is the scaled score or the raw score in absence of a scaling scheme. |
| [2] | The term sample genome means one genome which provides the main configuration for all individuals. |
| [3] | This GA uses non-overlapping populations. |
The Interactive Mode¶
Pyevolve have introduced the concept of the Interactive Mode in the course of evolution. When you are evolving, and the Interactive Mode is enabled, you can press the ESC Key anytime in the evolution process. By pressing that key, you will enter in the interactive mode, with a normal python prompt and the Interaction module exposed to you as the “it” module.
Warning
note that the Interactive Mode for Linux/Mac was disabled in the 0.6 release of Pyevolve. The cause was the platform dependant code. To use it in Linux/Mac you must set the generation in wich Pyevolve will enter in the Interactive Mode by using GSimpleGA.GSimpleGA.setInteractiveGeneration() method; see the Interaction module documentation for more information.
If you want to continue the evolution, just press CTRL-D on Linux or CTRL-Z on Windows.
See this session example:
# pyevolve_ex1_simple.py
Gen. 1 (0.20%): Max/Min/Avg Fitness(Raw) [6.18(11.00)/4.42(1.00)/5.15(5.15)]
Gen. 20 (4.00%): Max/Min/Avg Fitness(Raw) [11.70(15.00)/7.24(3.00)/9.75(9.75)]
Gen. 40 (8.00%): Max/Min/Avg Fitness(Raw) [17.99(21.00)/12.00(9.00)/14.99(14.99)]
Loading module pylab (matplotlib)... done!
Loading module numpy... done!
## Pyevolve v.0.6 - Interactive Mode ##
Press CTRL-D to quit interactive mode.
>>>
As you can see, when you press the ESC Key, a python prompt will be show and the evolution will be paused.
Now, what you can do with this prompt !?
- See all the current population individuals
- Change the individuals
- Plot graphics of the current population
- Data analysis, etc... python is your limit.
Note
to use graphical plots you will obviously need the Matplotlib, see more information in the Requirements section for more information.
Inspecting the population¶
This is a session example:
## Pyevolve v.0.6 - Interactive Mode ##
Press CTRL-Z to quit interactive mode.
>>> dir()
['__builtins__', 'ga_engine', 'it', 'population', 'pyevolve']
>>>
>>> population
- GPopulation
Population Size: 80
Sort Type: Scaled
Minimax Type: Maximize
Slot [Scale Method] (Count: 1)
Name: LinearScaling
Doc: Linear Scaling scheme
.. warning :: Linear Scaling is only for positive raw scores
- Statistics
Minimum raw score = 10.00
Minimum fitness = 13.18
Standard deviation of raw scores = 2.71
Maximum fitness = 19.92
Maximum raw score = 23.00
Fitness average = 16.60
Raw scores variance = 7.36
Average of raw scores = 16.60
>>> len(population)
80
>>> individual = population[0]
>>> individual
- GenomeBase
Score: 23.000000
Fitness: 19.920000
Slot [Evaluator] (Count: 1)
Name: eval_func
Slot [Initializator] (Count: 1)
Name: G1DListInitializatorInteger
Doc: Integer initialization function of G1DList
This initializator accepts the *rangemin* and *rangemax* genome parameters.
Slot [Mutator] (Count: 1)
Name: G1DListMutatorSwap
Doc: The mutator of G1DList, Swap Mutator
Slot [Crossover] (Count: 1)
Name: G1DListCrossoverSinglePoint
Doc: The crossover of G1DList, Single Point
.. warning:: You can't use this crossover method for lists with just one element.
- G1DList
List size: 50
List: [0, 5, 6, 7, 2, 0, 8, 6, 0, 0, 8, 7, 5, 6, 6, 0, 0, 3, 0, 4, 0, 0, 9, 0, 9, 2, 0, 0, 4, 2
, 5, 0, 0, 2, 0, 0, 0, 1, 8, 7, 0, 8, 9, 0, 8, 0, 0, 0, 9, 0]
The exposed modules and objects¶
The Interaction module is imported with the name “it”, you can see calling the python native dir():
>>> dir()
['__builtins__', 'ga_engine', 'it', 'population', 'pyevolve']
The namespace have the the following modules:
- ga_engine
- The GSimpleGA.GSimpleGA instance, the GA Engine.
- it
- The Interaction module, with the utilities and graph plotting functions.
- population
- The current population.
- pyevolve
- The main namespace, the pyevolve module.
Using the “it” module
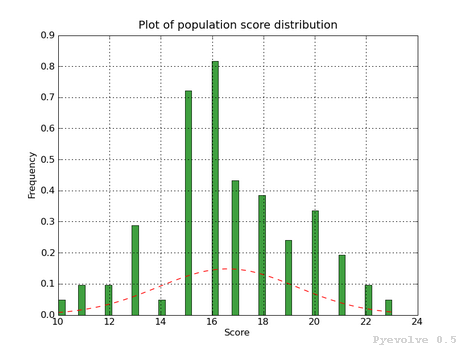
Plotting the current population raw scores histogram
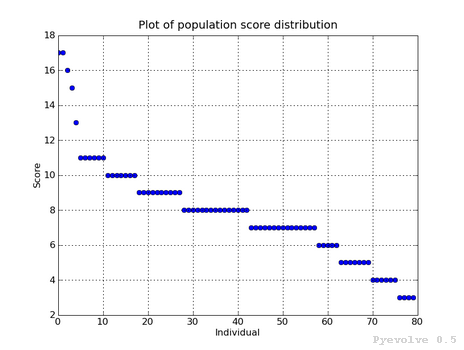
>>> it.plotHistPopScore(population)Plotting the current population raw scores distribution
>>> it.plotPopScore(population)Get all the population raw scores
>>> popScores = it.getPopScores(population) >>> popScores [17.0, 17.0, 16.0, 15.0, 13.0, 11.0, 11.0, 11.0, 11.0, 11.0, 11.0, 10.0, 10.0, 10.0, 10.0, 10.0, 10.0, 10.0, 9.0, 9.0, 9.0, 9.0, 9.0, 9.0, 9.0, 9.0, 9.0, 9.0, 8.0, 8.0, 8.0, 8.0, 8.0, 8.0, 8.0, 8.0, 8.0, 8.0, 8.0, 8.0, 8.0, 8.0, 8.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 7.0, 6.0, 6.0, 6.0, 6.0, 6.0, 5.0, 5.0 , 5.0, 5.0, 5.0, 5.0, 5.0, 4.0, 4.0, 4.0, 4.0, 4.0, 4.0, 3.0, 3.0, 3.0, 3.0]
Extending Pyevolve¶
Creating the representation¶
The first thing you must do is to see the source code of the GenomeBase.GenomeBase class or the source of the G1DBinaryString.G1DBinaryString class, they are very simple to understand.
Those are the steps to extend Pyevolve with a new representation:
- Create the chromosome representation class
- Create the initializator for your chromosome
- Create the genetic operators
- Create the Mutator
- Create the Crossover
Well, let’s begin with the creation of the elegant 1D Binary String chromosome, this chromosome is nothing more than a simple array of ‘0’s or ‘1’s, like that: “001001000”.
Note
This 1D Binary String chromosome is an Pyevolve existing chromosome, of course, in the Pyevolve implementation we have more features that we will implement here in this simple example.
All of our new chromosomes must extend the base class called GenomeBase.GenomeBase, this class contains the basic slots for the genetic operators and all the internal stuff that you don’t need to care.
Let’s code the initial draft of our chromosome class:
from GenomeBase import GenomeBase
class G1DBinaryString(GenomeBase):
pass
As you see, we have imported the GenomeBase.GenomeBase class from the GenomeBase module and we have created the G1DBinaryString class extending the base class.
The next step is to create our constructor method for our class, I’ll show it before and explain later:
def __init__(self, length=10):
GenomeBase.__init__(self)
self.genomeString = []
self.stringLength = length
self.initializator.set(Consts.CDefG1DBinaryStringInit)
self.mutator.set(Consts.CDefG1DBinaryStringMutator)
self.crossover.set(Consts.CDefG1DBinaryStringCrossover)
Well, we start by calling the base class constructor and then creating an internal list to hold our ‘0’s and ‘1’s. It is important to note that we don’t initialize the list, this will be done by our initializator function, and it is because of this that we must keep as an internal attribute the length of your 1D Binary String.
Next, we set our initializator, mutator and crossover to constants, this constants have just the functions of our genetic operators, but if you want, you can set they later, in this example, we will use the defaults for the G1D Binary String.
Note
The attributes self.initializator, self.mutator and self.crossover are all inherited from the GenomeBase class. They are all function slots (FunctionSlot.FunctionSlot).
Now, you must provide the copy() and clone() methods for your chromosome, because they are used to replicate the chromosome over the population or when needed by some genetic operators like reproduction.
The copy() method is very simple, what you need to do is to create a method that copy the contents of your chromosome to another chromosome of the G1DBinaryString class.
Here is our copy() method:
def copy(self, g):
""" Copy genome to 'g' """
GenomeBase.copy(self, g)
g.stringLength = self.stringLength
g.genomeString = self.genomeString[:]
As you can see, we first call the base class copy() method and later we copy our string length attribute and our internal genomeString, which is our list of ‘0’s and ‘1’s.
Warning
It is very important to note that you must COPY and not just create a reference to the object. On the line that we have the self.genomeString[:], if you use just self.genomeString, you will create a REFERENCE to this object and not a copy. This a simple warning, but can avoid many headaches.
The next step is to create our clone() method, the clone method, as the name says, is a method which return another instance of the current chromosome with the same contents.
Let’s code it:
def clone(self):
""" Return a new instace copy of the genome """
newcopy = G1DBinaryString(self.stringLength)
self.copy(newcopy)
return newcopy
We simple create a new instance and use the copy() method that we have created to copy the instance contents.
Ready ! We have our first representation chromosome. You can add many more features by implementing python operators like __getitem__, __setitem__.
Creating the initializator¶
Sorry, not written yet.
Creating the mutator¶
Sorry, not written yet.
Creating the crossover¶
The file Crossovers.py implements the crossover methods available in Pyevolve. So, that is where you should look to implement your new crossover method. The process of adding a new crossover method is as follows:
- Create a new method such that the name reflects the type of chromosome representation it works with, and the crossover method name. For example, Crossovers.G1DListCrossoverRealSBX(), can work with 1D List representations and it operates on real values and it is the SBX crossover operator.
- The method must take two parameters, ‘genome’ and ‘args’.
- From ‘args’, get the two parents which will take part in the crossover, gMom and gDad.
- Once you have gMom and gDad, use them to create the two children, sister and brother.
- Simply return the sister and brother.
Any constants that your crossover method uses should be defined in Consts.py (Consts).
Genetic Programming Tutorial¶
In the release 0.6 of Pyevolve, the new Genetic Programming core was added to the framework. In the Example 18 - The Genetic Programming example you’ll see how simple and easy is Pyevolve GP core when compared with other static-typed languages.
Here is a simple example:
from pyevolve import Util
from pyevolve import GTree
from pyevolve import GSimpleGA
from pyevolve import Consts
import math
rmse_accum = Util.ErrorAccumulator()
def gp_add(a, b): return a+b
def gp_sub(a, b): return a-b
def gp_mul(a, b): return a*b
def gp_sqrt(a): return math.sqrt(abs(a))
def eval_func(chromosome):
global rmse_accum
rmse_accum.reset()
code_comp = chromosome.getCompiledCode()
for a in xrange(0, 5):
for b in xrange(0, 5):
evaluated = eval(code_comp)
target = math.sqrt((a*a)+(b*b))
rmse_accum += (target, evaluated)
return rmse_accum.getRMSE()
def main_run():
genome = GTree.GTreeGP()
genome.setParams(max_depth=4, method="ramped")
genome.evaluator += eval_func
ga = GSimpleGA.GSimpleGA(genome)
ga.setParams(gp_terminals = ['a', 'b'],
gp_function_prefix = "gp")
ga.setMinimax(Consts.minimaxType["minimize"])
ga.setGenerations(50)
ga.setCrossoverRate(1.0)
ga.setMutationRate(0.25)
ga.setPopulationSize(800)
ga(freq_stats=10)
best = ga.bestIndividual()
print best
if __name__ == "__main__":
main_run()
Let’s work now step by step on the code to learn what each building block means, the first part you see the imports:
from pyevolve import Util
from pyevolve import GTree
from pyevolve import GSimpleGA
from pyevolve import Consts
import math
In the Util module is where we’ll found many utilities functions and classes like Util.ErrorAccumulator. The GTree is where resides the GTree.GTreeGP class, which is the main genome used by the GP core of Pyevolve. Note that we are importing the GSimpleGA module, in fact, the GA core will detect when you use a Genetic Programming genome and will act as the GP core. The modules Consts and math imported here are for auxiliary use only. Next we have:
rmse_accum = Util.ErrorAccumulator()
Here we instantiate the Util.ErrorAccumulator, which is a simple accumulator for errors. It has methods for getting Adjusted Fitness, Mean Square Error, Root Mean Square Error, mean, squared or non-squared error measures. In the next block we define some GP operators:
def gp_add(a, b): return a+b
def gp_sub(a, b): return a-b
def gp_mul(a, b): return a*b
def gp_sqrt(a): return math.sqrt(abs(a))
See that they are simple Python functions starting with the “gp” prefix, this is important if you want that Pyevolve automatically add them as non-terminals of the GP core. As you can note, the square root is a protected square root, since it uses the absolute value of “a” (we don’t have square root of negative numbers, except in the complex analysis). You can define any other function you want. Later we have the declaration of the Evaluation function for the GP core:
def eval_func(chromosome):
global rmse_accum
rmse_accum.reset()
code_comp = chromosome.getCompiledCode()
for a in xrange(0, 5):
for b in xrange(0, 5):
evaluated = eval(code_comp)
target = math.sqrt((a*a)+(b*b))
rmse_accum += (target, evaluated)
return rmse_accum.getRMSE()
As you see, the eval_func() receives one parameter, the chromosome (the GP Tree in our case, an instance of the GTree.GTreeGP class). We first declare the global error accumulator and reset it, since we’ll start to evaluate a new individual, a new “program”. In the line where we call GTree.GTreeGP.getCompiledCode(), here is what happens: Pyevolve will get the pre ordered expression of the GP Tree and then will compile it into Python bytecode, and will return to you an object of the type “code”. This object can then be executed using the Python native eval() function. Why compiling it in bytecodes ? Because if we don’t compile the program into Python bytecode, we will need to parse the Tree every time we want to evaluate our program using defined variables, and since this is a commom use of the GP program, this is the fastest way we can do it in pure Python.
The next block we simple iterate using two variables “a” and “b”.
Note
Please note that the variable names here is the same that we will use as terminals later.
What you see now is the evaluation of the “code_comp” (which is the GP individual) and the evaluation of the objective function in which we want to fit (the Pythagorean theorem). Next we simple add the “target” value we got from the Pythagorean theorem and the “evaluated” value of the individual to the Error Accumulator. In the end of the evaluation function, we return the Root Mean Square Error. If you don’t like to add the evaluated and the target values using a tuple, you can use the Util.ErrorAccumulator.append() method, which will give the same results.
Next we start to define our main_run() function:
def main_run():
genome = GTree.GTreeGP()
genome.setParams(max_depth=4, method="ramped")
genome.evaluator.set(eval_func)
The first thing we instantiate here is the GTree.GTreeGP class (the GP individual, the Tree). Next we set some parameters of the GTreeGP. The first is the “max_depth”, which is used by genetic operators and initializators to control bloat, in this case, we use 4, which means that no Tree with a height > 4 will grow. Next we set the “method”, this is the initialization method, and the values accepted here depends of the initialization method used, since we do not have specified the initialization method, Pyevolve will use the default, which is the Initializators.GTreeGPInitializator() (it accepts “grow”, “full” and “ramped” methods for Tree initialization. And in the last line of this block, we set the previously defined evaluation function called eval_func(). In the next block we then instantiate the GSimpleGA core and set some parameters:
ga = GSimpleGA.GSimpleGA(genome)
ga.setParams(gp_terminals = ['a', 'b'],
gp_function_prefix = "gp")
The “ga” object will hold an instance of the GSimpleGA.GSimpleGA class, which is the core for both Genetic Algorithms and Genetic Programming. Pyevolve will automatically detect if you are creating a environment for a GP or for a GA. Next we set some parameters of the core, the first is a list called “gp_terminals”. The “gp_terminals” will hold the “variables” or in GP vocabulary . Note that the name of the terminals are the same we used in our evaluation function called eval_func(). The next step is to define the prefix of the GP operators (functions) or the Non-terminal node. Pyevolve will automatically search for all functions defined in the module which starts with “gp” (example: gp_sub, gp_add, gp_IHateJava, etc...) and will add these functions as the non-terminal nodes of the GP core.
The next part of the code is almost the same as used in the Genetic Algorithms applications, they are the EA parameters to setup and start the evolution:
ga.setMinimax(Consts.minimaxType["minimize"])
ga.setGenerations(50)
ga.setCrossoverRate(1.0)
ga.setMutationRate(0.25)
ga.setPopulationSize(800)
ga(freq_stats=10)
best = ga.bestIndividual()
print best
And in the last part of the source code, we have:
if __name__ == "__main__":
main_run()
This part is important, since Pyevolve needs to know some information about objects in the main module using instrospection, you MUST NEED to declare this checking, the multiprocessing module of Python only works with this too, so if you’re planning to use it, please do not forget.
And that’s it, you have done your first GP program.
Visualizing individuals¶
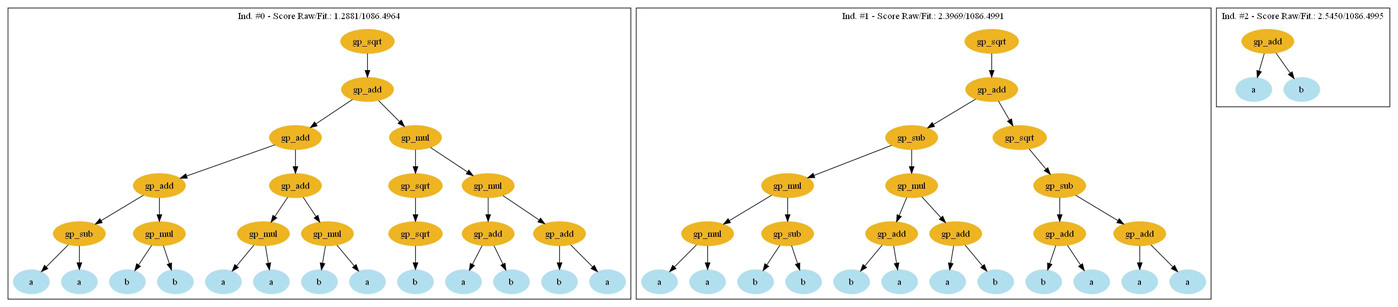
Pyevolve comes with a plotting utility to make pictures of your GP individuals, it uses the “pydot” and “Graphviz” to create those images. See more information in the Requirements section. What you need to change to see, for example, the first 3 best individuals of your first generation is to add a Step callback function into the code, let’s first define the callback function:
def step_callback(gp_engine):
if gp_engine.getCurrentGeneration() == 0:
GTree.GTreeGP.writePopulationDot(gp_engine, "trees.jpg", start=0, end=3)
The code is self explanative, the parameter is the GP core, first we check if it is the first generation and then we use the GTree.GTreeGP.writePopulationDot() method to write to the “trees.jpg” file, the range from 0 and 3 individuals of the population. Then in the main function where we instantiate the GP core, we simple use:
ga.stepCallback.set(step_callback)
And the result will be:
Snippets¶
Here are some snippets to help you.
Using two mutators at same time¶
To use two mutators at same time, you simple add one more to the mutator function slot, like this:
>>> genome.mutator.set(Mutators.G1DListMutatorRealGaussian) >>> genome.mutator.add(Mutators.G1DListMutatorSwap)The first line will set the Mutators.G1DListMutatorRealGaussian(), and the second line add one more mutator, the Mutators.G1DListMutatorSwap().
As you can see, it’s very simple and easy, and you will have two mutation operators at same time.
If you want, that just one of this mutators (random picked) be executed at the mutation process, set the random apply parameter of the FunctionSlot.FunctionSlot class to True
>>> genome.mutator.setRandomApply(true)
Using one allele to all list (chromosome) elements (genes)¶
Sometimes you want to use just one allele type to all genes on the 1D List or other chromosomes, you simple add one allele type and enable the homogeneous flag to True:
>>> setOfAlleles = GAllele.GAlleles(homogeneous=True) >>> lst = [ "1", "two", 0, 777 ] >>> a = GAllele.GAlleleList(lst) >>> setOfAlleles.add(a)Ready, your setOfAlleles is the GAllele.GAlleles class instance with the lst ([ “1”, “two”, 0, 777 ]) as alleles in all genes.
Changing the selection method¶
To change the default selection method, you must do this:
>>> ga = GSimpleGA.GSimpleGA(genome) >>> ga.selector.set(Selectors.GTournamentSelector)In this example, we are changing the selection method to the Selectors.GTournamentSelector(), the Tournament Selector.
Doing the same evolution on with random seed¶
Using a random seed, you can guarantee that the evolution will be always the same, no matter the number of executions you make. To initialize the GA Engine with the random seed, use the seed parameter when instantiating the GSimpleGA.GSimpleGA class:
ga_engine = GSimpleGA(genome, 123) # or ga_engine = GSimpleGA(genome, seed=123)The value 123 will be passed as the random seed of the GA Engine.
Writing the evolution statistics to a CSV File¶
You can write all the statistics of an evolution to a CSV (Comma Separated Values) fil using the DB Adapter called DBAdapters.DBFileCSV, just create an instance of the adapter and attach it to the GA Engine:
csv_adapter = DBFileCSV(identify="run1", filename="stats.csv") ga_engine.setDBAdapter(csv_adapter)Ready ! Now, when you run your GA, all the stats will be dumped to the CSV file. You can set the frequency in which the stats will be dumped, just use the parameter frequency of the DBFileCSV.
Use the HTTP Post to dump GA statistics¶
With the DBAdapters.DBURLPost, you can call an URL with the population statistics in every generation or at specific generation of the evolution:
urlpost_adapter = DBURLPost("http://localhost/post.py", identify="run1", frequency=100) ga_engine.setDBAdapter(urlpost_adapter)Now, the URL “http://localhost/post.py” will be called with the statistics params in every 100 generations. By default, the adapter will use the HTTP POST method to send the parameters, but you can use GET method setting the post paramter to False.
See the mod:Statistics and DBAdapters.DBURLPost documentation.
Using two or more evaluation function¶
To use two or more evaluation function, you can just add all the evaluators to the slot:
genome.evaluator.set(eval_func1) genome.evaluator.add(eval_func2)The result raw score of the genome, when evaluated using more then on evaluation function, will be the sum of all returned scores.
Note
the method set of the function slot remove all previous functions added to the slot.
Real-time statistics visualization¶
You have three options to view the statistics while in the course of the evolution:
Console statistics
You can view the statistics by setting the freq_stats parameter of the GSimpleGA.GSimpleGA.evolve() method. It will dump the statistics in the console.Using the sqlite3 DB Adapter
You can use the DBAdapters.DBSQLite DB Adapter and set the commit_freq to a low value, so you can use the Graphical Plotting Tool of Pyevolve to create graphics while evolving.Using the VPython DB Adapter
Use the DBAdapters.DBVPythonGraph DB Adapter, this DB Adapter will show four statistical graphs, it is fast and easy to use.
How to manually add non-terminal functions to Genetic Programming core¶
When you set Pyevolve to automatically catch non-terminal funcions for your GP core you do something like this:
ga = GSimpleGA.GSimpleGA(genome)
ga.setParams(gp_terminals = ['a', 'b'],
gp_function_prefix = "gp")
The “gp_function_prefix” paremter tells Pyevolve to catch any function starting with “gp”. But there are times that you want to add each function manually, so you just need to add a dictionar parameter called “gp_function_set”, like this:
ga.setParams(gp_terminals = ['a', 'b'],
gp_function_set = {"gp_add" :2,
"gp_sub" :2,
"gp_sqrt":1})
Note the “gp_function_set” dictionary parameter which holds as key the function name and for the value, the number of arguments from that function, in this case we have “gp_add” with 2 parameters, “gp_sub” with 2 and “gp_sqrt” with just one.
Passing extra parameters to the individual¶
Sometimes we want to add extra parameters which we need the individuals must carry, in this case, we can use the method GenomeBase.GenomeBase.setParams() to set internal parameters of the individual and the method GenomeBase.GenomeBase.getParam() to get it’s parameters back, see an example:
def evaluation_function(genome):
parameter_a = genome.getParam("parameter_a")
def main():
# (...)
genome = G1DList.G1DList(20)
genome.setParams(rangemin=-5.2, rangemax=5.30, parameter_a="my_value")
# (...)
Note
Due to performance issues, Pyevolve doesn’t copy the internal parameters into each new created individual, it simple references the original parameters, this reduces memory and increases speed.
Using ephemeral constants in Genetic Programming¶
You can use an ephemeral constant in Pyevolve GP core by using the “ephemeral:” prefix in your GP terminals, like in:
ga = GSimpleGA.GSimpleGA(genome)
ga.setParams(gp_terminals = ['a', 'b', 'ephemeral:random.randint(1,10)'],
gp_function_prefix = "gp")
In this example, the ephemeral constant will be an integer value between 1 and 10. You can use any method of the Python random module to specify the ephemeral constant.